The input file can have no, one or more rows of column labels and no, one or more columns of row labels. The sample input file shown below has 1 row of column labels and 2 columns of row labels.
| rc1 | rc2 | col1 | col2 | col3 |
| rowLabel11 | rowLabel12 | 1 | 2 | 3 |
| rowLabel21 | rowLabel22 | 2 | 1 | 2 |
| rowLabel31 | rowLabel32 | 3 | 2 | 3 |
| rowLabel41 | rowLabel42 | 3 | 1 | 2 |
| rowLabel51 | rowLabel52 | 1 | 2 | 2 |
| rowLabel61 | rowLabel62 | 3 | 1 | 3 |
| rowLabel71 | rowLabel72 | 1 | 2 | 3 |
| rowLabel81 | rowLabel82 | 3 | 1 | 2 |
| rowLabel91 | rowLabel92 | 1 | 2 | 3 |
The LAS wizard guides you through the various settings of the algorithm. Let me thank to Manish Ranjan Kumar for the C# Wizard Control, which I found very helpful.
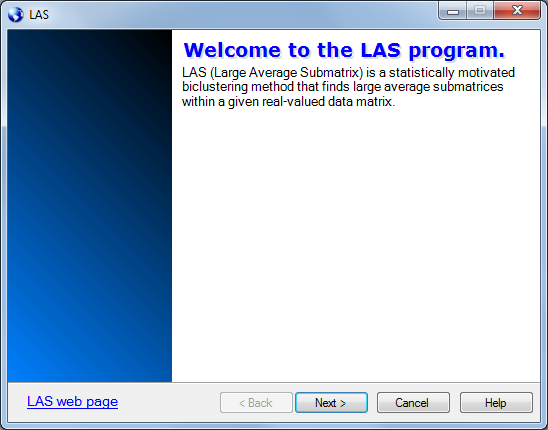
The second page of the wizard helps you load the data to be biclustered.
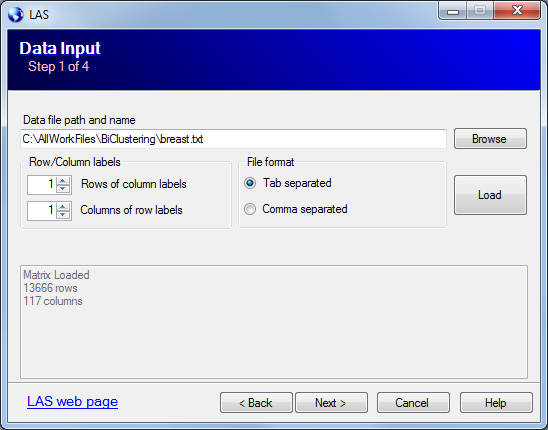
The third page of the wizard controls the data normalization.
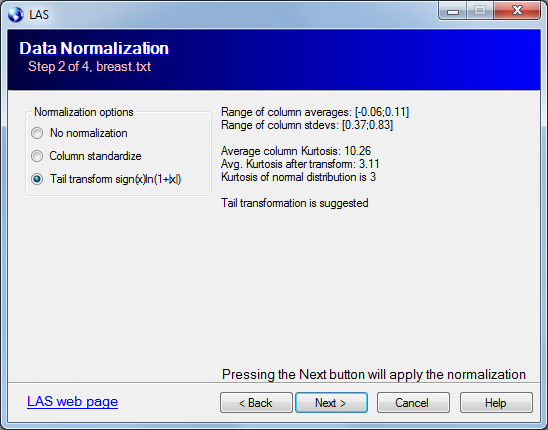
The LAS model assumes the noise component of the data to have i.i.d. N(0,1) distribution. To make the data closer the model we can apply one of the normalization:
The program calculated the average column Kurtosis after second 2 normalizations and suggests the one with Kurtosis closer to 3.
The forth page of the wizard controls the parameters of biclustering.

"Search for biclusters with" option sets whether we are interested in biclusters with only large positive average, large negative average, or both.
The "Stopping Criteria" defines the maximum number of biclusters to produce and the "Score cut off". Once a bicluster with the score lower than the cut off is produced the search is stopped.
Each produced bicluster is the best among the "Number of iterations of the search algorithm per bicluster" found by the search procedure. The "Number of threads to use" sets the number of processor cores used by the program. To run LAS in background set it to at most 1 less than the maximum value (number of cores for your machine).
The last page of the wizard shows the progress of the bicluster search and controls the location and the type of output files.
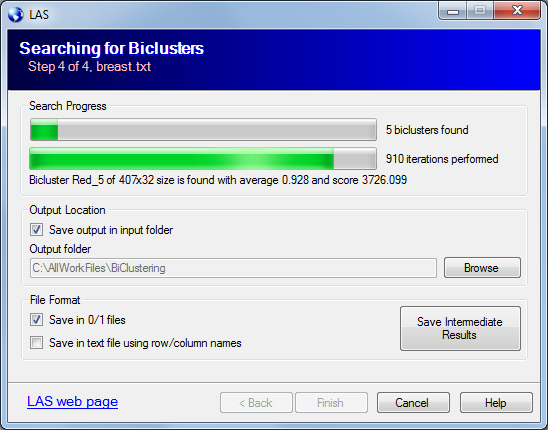
The "Search Progress" shows the number of biclusters found so far. The checkbox "Save output in input folder" will fill the "Output folder" field for you with the path to the input file.
You do not have to wait for all biclusters to be found, the "Save Intermediate Results" button allows you to save the biclusters found up to the current moment.
There are two formats for the output files: Binary (0/1) and Text. The names of output files are formed from the name of the input file:
Let the name of the input file be:
The Binary output consists of 3 files:
The text output consists of 3 files:
The common file "..Labels.txt" contains the summary information about biclusters. Each row contains information about a single bicluster, i.e. its size, average and score. For the breast cancer data it looks like this:
| Red_1 | bicluster of | 1431 | x | 43 | size with average | 0.7551 | and score | 12894.1 |
| Red_2 | bicluster of | 1520 | x | 29 | size with average | 0.8491 | and score | 11067.5 |
| Red_3 | bicluster of | 1486 | x | 26 | size with average | 0.8061 | and score | 7803.8 |
| … | … | … | … | … | … | … | … | … |
The binary output files "..Rows.binary.txt" and "..Columns.binary.txt" contain information about biclusters' row- and column-sets respectively. For a data matrix with m rows and n columns, each row of the "..Rows.binary.txt" file will contain m 0/1 values and each row of the "..Columns.binary.txt" file will contain n 0/1 values.
Let's consider a data matrix with 4 rows and 5 columns with the following output files.
| ..Rows.binary.txt file | ..Columns.binary.txt file | ||||||||
| 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |
| 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | |
The first row in both files keeps the information about the first bicluster (matching the first row in ..Labels.txt file). The rows and columns of the bicluster are indicated by 1s. So the first bicluster contains the 2nd and 3rd rows and 1st, 3rd and 4th columns:
| The data matrix with first bicluster: | |||||
| col1 | col2 | col3 | col4 | col5 | |
| row1 | 4 | 2 | 3 | 1 | 0 |
| row2 | 6 | 7 | 5 | 9 | 4 |
| row3 | 9 | 1 | 8 | 9 | 1 |
| row4 | 0 | 2 | 4 | 2 | 1 |
As the binary output files, the text output files contain information about biclusters' rows ("..Rows.text.txt") and columns ("..Columns.text.txt"), one bicluster per row in each file. Namely, the kth row in "..Rows.text.txt" file contains labels of all rows of the kth bicluster and the kth row in "..Columns.text.txt" file contains labels of all columns of the kth bicluster. The biclusters described above by the binary files will be represented in the text files in the following way:
| ..Rows.text.txt file | ..Columns.text.txt file | ||||
| row2 | row3 | col1 | col3 | col4 | |
| row1 | row3 | col4 | col5 | ||